This chapter may be moved from the printed PDF copy to an online Appendix.
For ease of exposition, the chapters in Part IV of this book used small examples to illustrate the statistical and visualization aspects of the techniques as they were described. Some of the main examples were spread over Chapters 10–13 to illustrate the specific statistical and graphical methods of those chapters.
Here I present more complete analyses of a few datasets to place the methods of this book in the wider research context. I illustrate a sequence of steps, starting with research questions involved and proceeding to the graphical, computing and statistical methods of this book required to understand and explain these data.1
Packages
In this chapter I use the following packages. Load them now.
14.2 Bivariate views
Corrgram
With this many variables, it is useful to view the correlations among them in a corrgram (sec-corrgram), giving a high-level reconnaissance. As noted earlier, the corrgram::corrgram() function can enhance perception by permuting the variables in the order of their variable vectors in a biplot, so more highly correlated variables are adjacent in the plot, an example of effect ordering for data displays (Friendly & Kwan, 2003).
NeuroCog |>
select(-Dx, -SocialCog) |>
corrgram(order = TRUE,
diag.panel = panel.density,
upper.panel = panel.pie)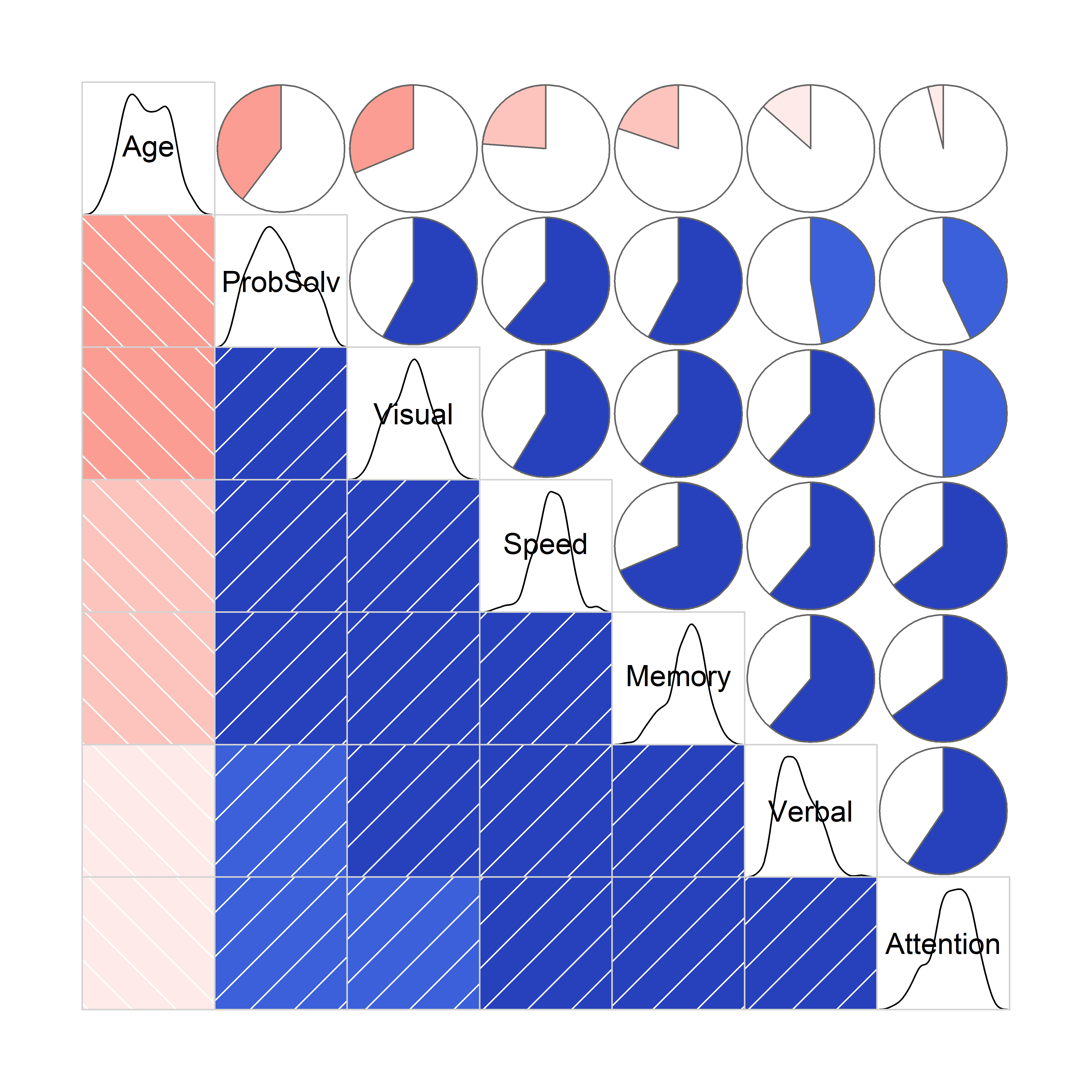
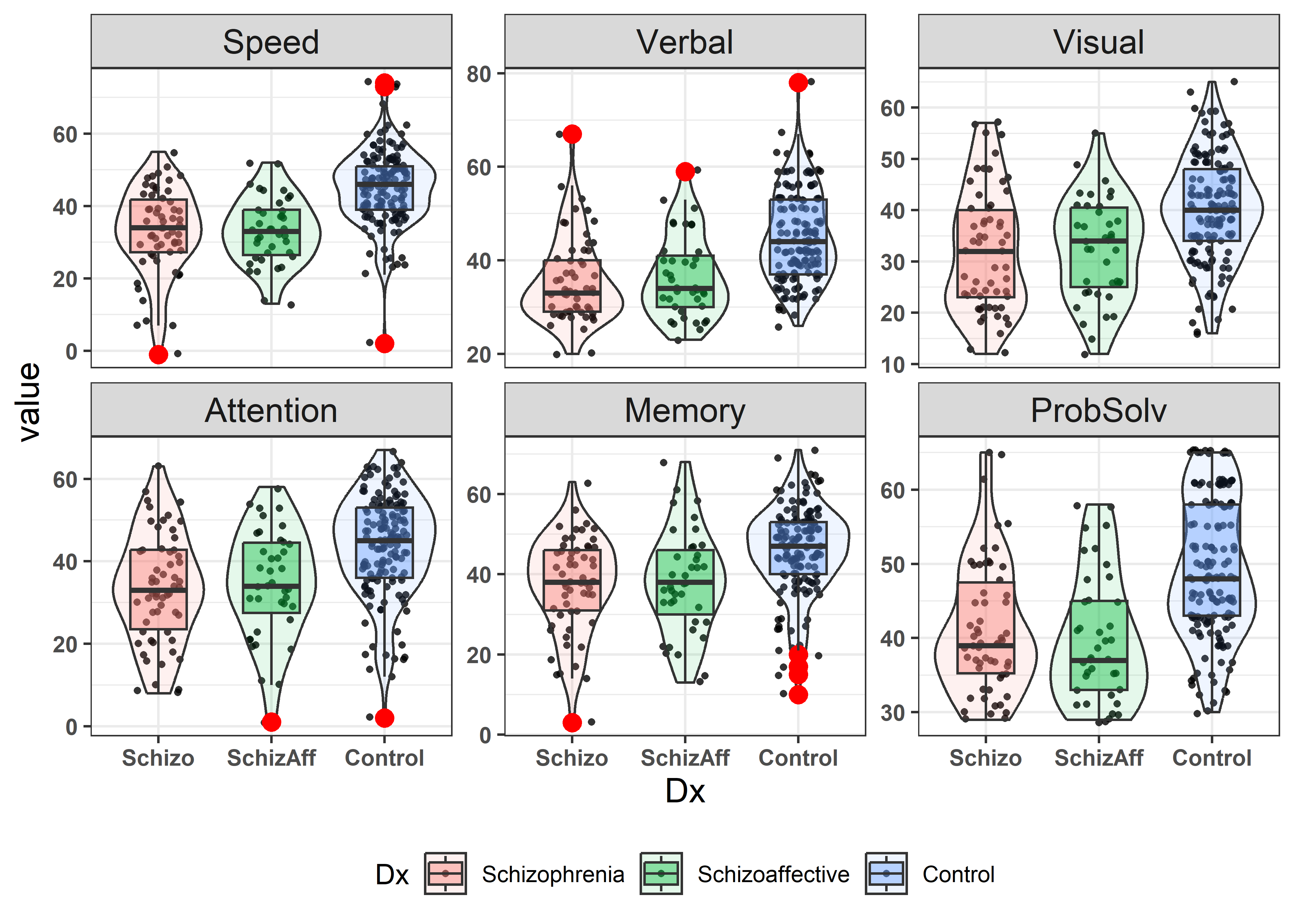
NeuroCog data. The upper and lower triangles use two different ways of encoding the value of the correlation for each pair of variables.
In this plot you can see that adjacent variables are more highly correlated than those more widely separated. The diagonal panels show that most variables are reasonably symmetric in their distributions. Age, not included in this analysis is negatively correlated with the others: older participants tend to do less well on these tests.
Scatterplot matrix
A scatterplot matrix gives a more detailed overview of all pairwise relations. The plot below suppresses the data points and summarizes the relation using data ellipses and regression lines. The model syntax, ~ Speed + ... |Dx, treats Dx as a conditioning variable (similar to the use of the color aestheic in ggplot2) giving a separate data ellipse and regression line for each group. (The legend is suppressed here. The groups are: Schizophrenic, SchizoAffective, Normal.)
col <- c("red", "darkgreen", "blue")
scatterplotMatrix(~ Speed + Attention + Memory + Verbal + Visual + ProbSolv | Dx,
data=NeuroCog,
plot.points = FALSE,
smooth = FALSE,
legend = FALSE,
col = col,
ellipse=list(levels=0.68))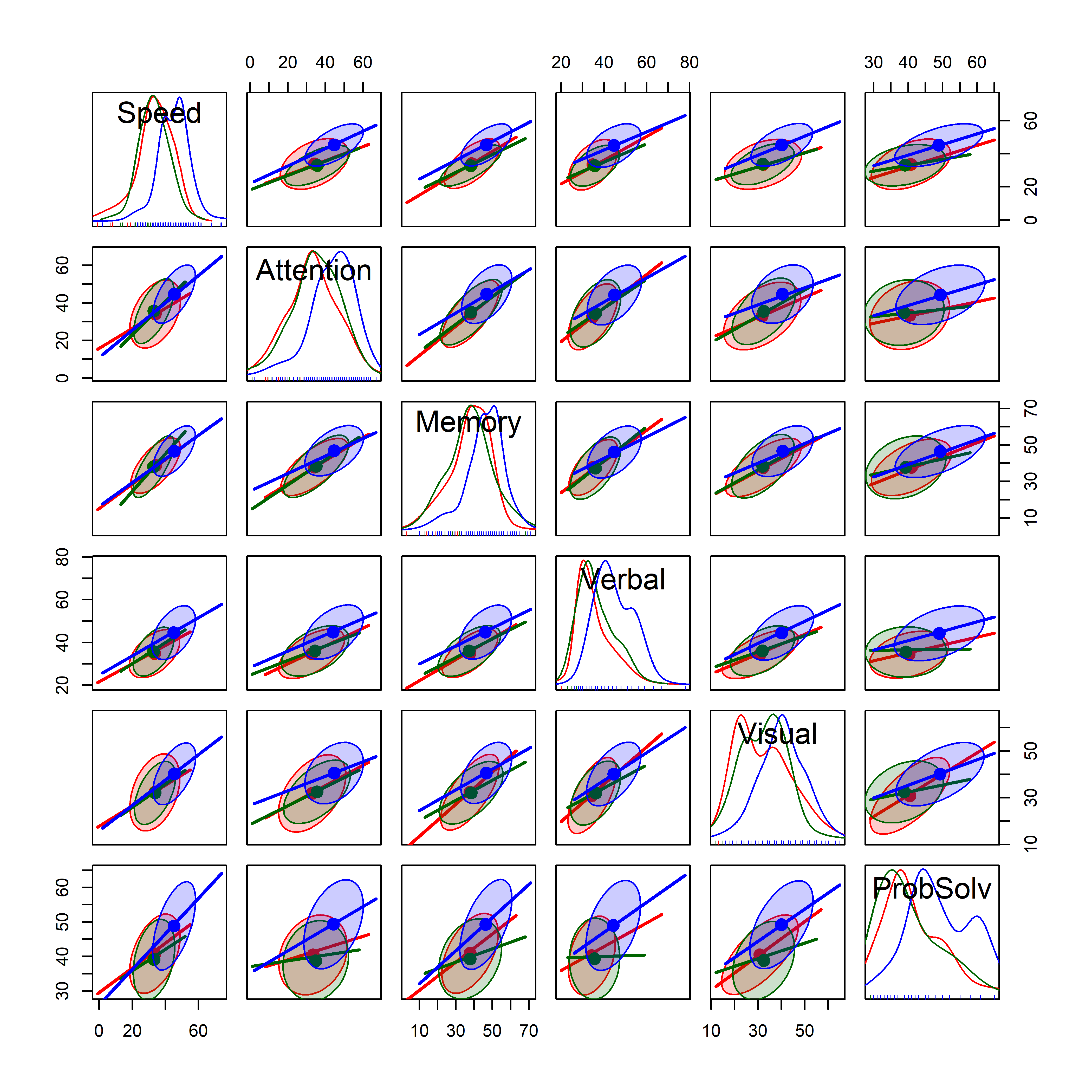
NeuroCog data. Points are suppressed here, focusing on the data ellipses and regression lines. Colors for the groups: Schizophrenic SchizoAffective Control
In Figure fig-NC-scatmat, you can see that the regression lines have similar slopes and similar data ellipses for the groups, though with a few exceptions. You can also see that the Normal group has higher means on all the variables and that the Schizophrenic and SchizoAffective don’t seem to differ very much.
Biplot
While still in exploratory mode, we can gain greater insight with our trusty multivariate juicer, the biplot (sec-biplot). A PCA of the cognitive variables shows that nearly 80% of the total variance is accounted for by two dimensions, of which the first dimension accounts for 69%.
neuro.pca <- NeuroCog |>
select(Speed:Visual) |>
prcomp(scale. = TRUE)
summary(neuro.pca)
# Importance of components:
# PC1 PC2 PC3 PC4 PC5
# Standard deviation 1.856 0.720 0.629 0.5761 0.5570
# Proportion of Variance 0.689 0.104 0.079 0.0664 0.0621
# Cumulative Proportion 0.689 0.793 0.872 0.9379 1.0000The biplot for this analysis (Figure fig-neuro-biplot) provides a nice summary of what was seen in the scatterplot matrix (Figure fig-NC-scatmat): A large difference between the Control group and the others, which don’t differ much from each other.
ggbiplot(neuro.pca,
obs.scale = 1, var.scale = 1,
groups = NeuroCog$Dx,
varname.size = 5,
var.factor = 1.5,
ellipse = TRUE) +
scale_color_discrete(name = 'groups') +
theme_minimal() +
theme(legend.direction = 'horizontal',
legend.position = 'top')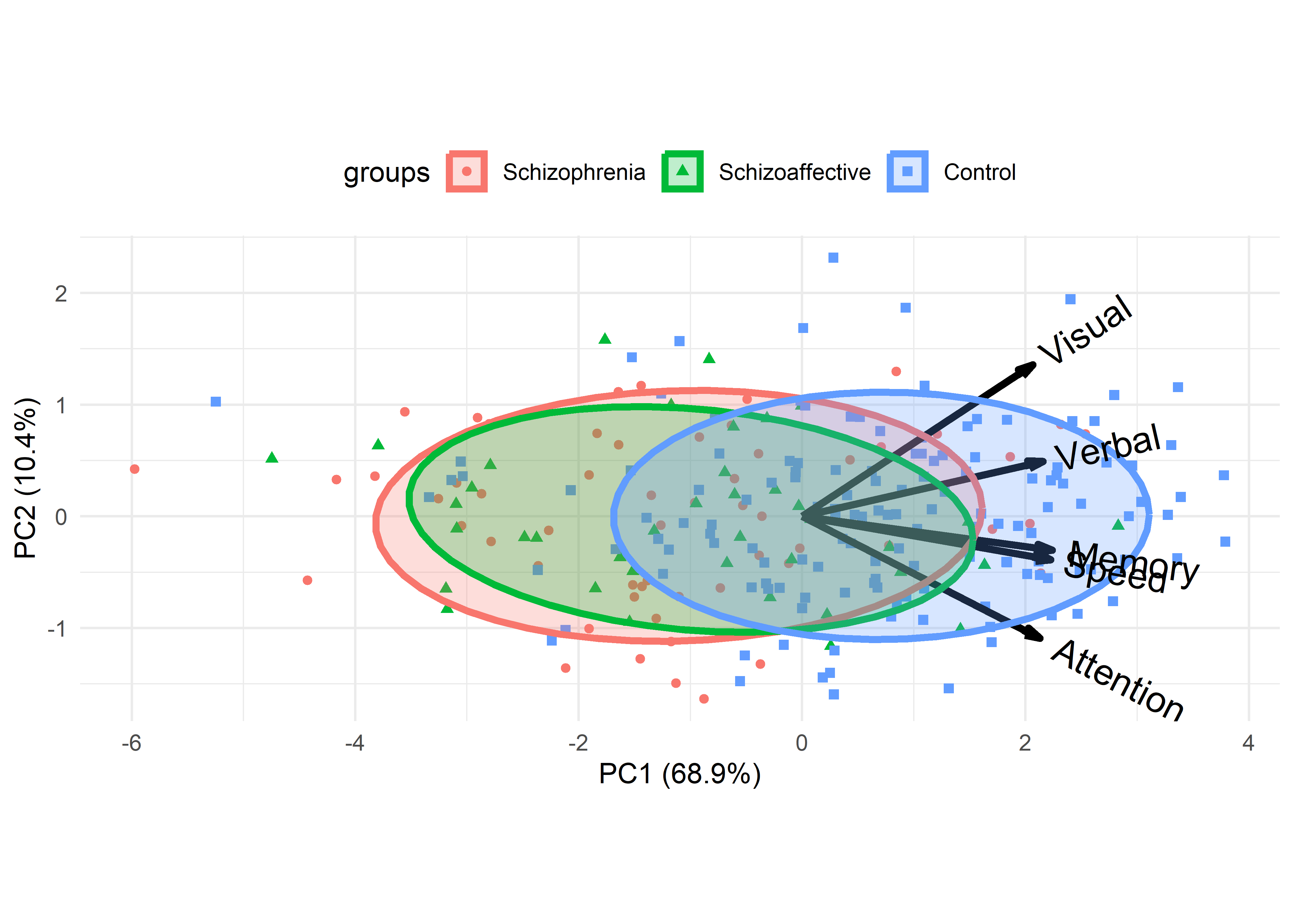
The variable vectors in Figure fig-neuro-biplot reflect the correlations of the response variables with each other and with the principal components, PC1 and PC2. All of the variables are positively correlated in this 2D view, Memory and Speed most highly so.
14.3 Fitting the MLM
We proceed to fit the one-way MANOVA model to answer the main research question of how well these variables distinguish among the groups.
NC.mlm <- lm(cbind(Speed, Attention, Memory, Verbal, Visual, ProbSolv) ~ Dx,
data=NeuroCog)
Anova(NC.mlm)
#
# Type II MANOVA Tests: Pillai test statistic
# Df test stat approx F num Df den Df Pr(>F)
# Dx 2 0.299 6.89 12 470 1.6e-11 ***
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1The first research question is captured by the contrasts for the Dx factor shown above. We can test these with car::linearHypothesis(). The contrast Dx1 for control vs. the diagnosed groups is highly significant,
# control vs. patients
print(linearHypothesis(NC.mlm, "Dx1"), SSP=FALSE)
#
# Multivariate Tests:
# Df test stat approx F num Df den Df Pr(>F)
# Pillai 1 0.289 15.9 6 234 2.8e-15 ***
# Wilks 1 0.711 15.9 6 234 2.8e-15 ***
# Hotelling-Lawley 1 0.407 15.9 6 234 2.8e-15 ***
# Roy 1 0.407 15.9 6 234 2.8e-15 ***
# ---
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1However, the second contrast, Dx2, comparing the schizophrenic and schizoaffective group, shows no difference in the collection of response means.
# Schizo vs SchizAff
print(linearHypothesis(NC.mlm, "Dx2"), SSP=FALSE)
#
# Multivariate Tests:
# Df test stat approx F num Df den Df Pr(>F)
# Pillai 1 0.006 0.249 6 234 0.96
# Wilks 1 0.994 0.249 6 234 0.96
# Hotelling-Lawley 1 0.006 0.249 6 234 0.96
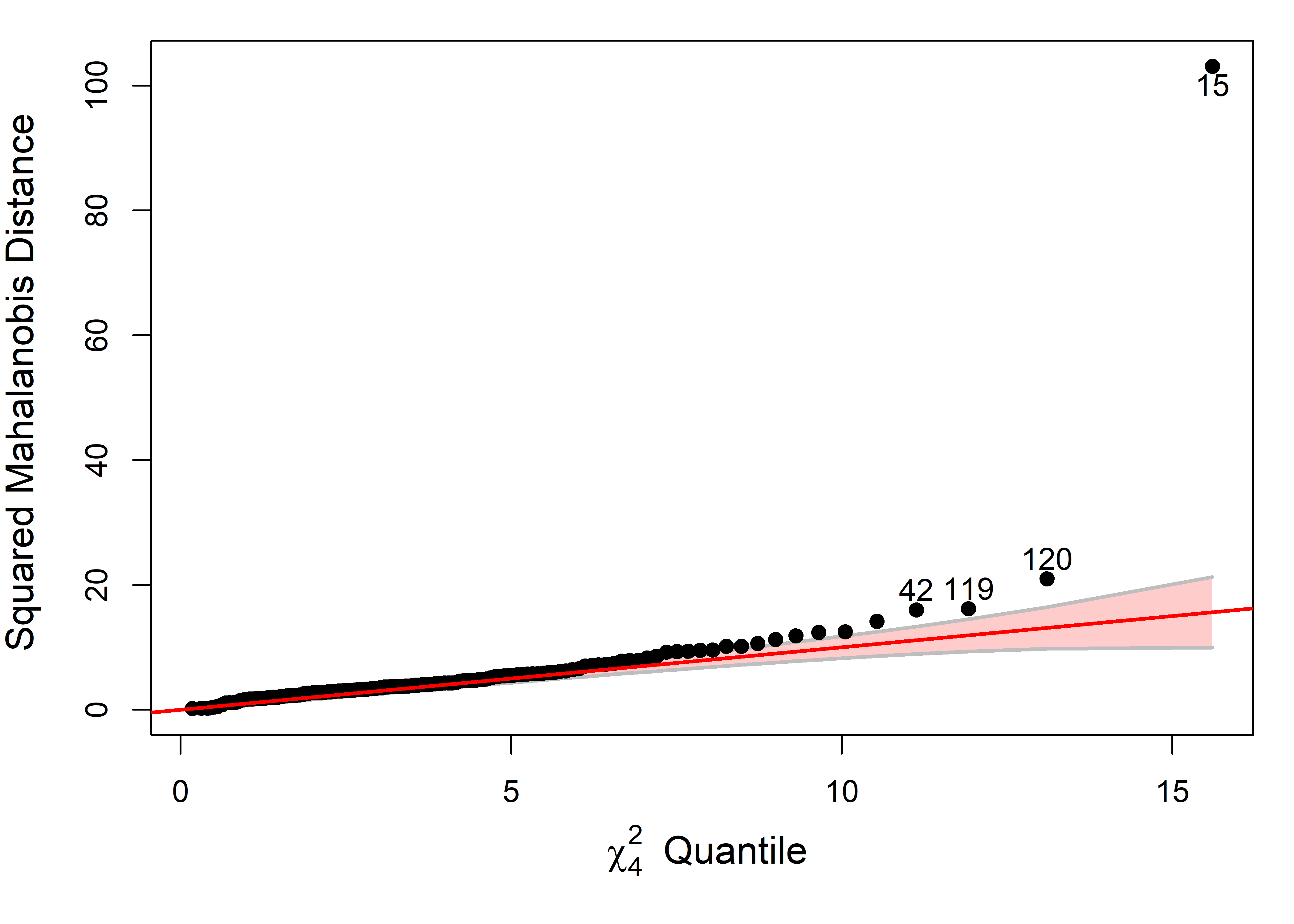
# Roy 1 0.006 0.249 6 234 0.96As a quick check on the model, a \(\chi^2\) QQ plot (Figure fig-NC-cqplot) reveals no problems with multivariate normality of residuals nor potentially harmful residuals.
cqplot(NC.mlm, id.n = 3)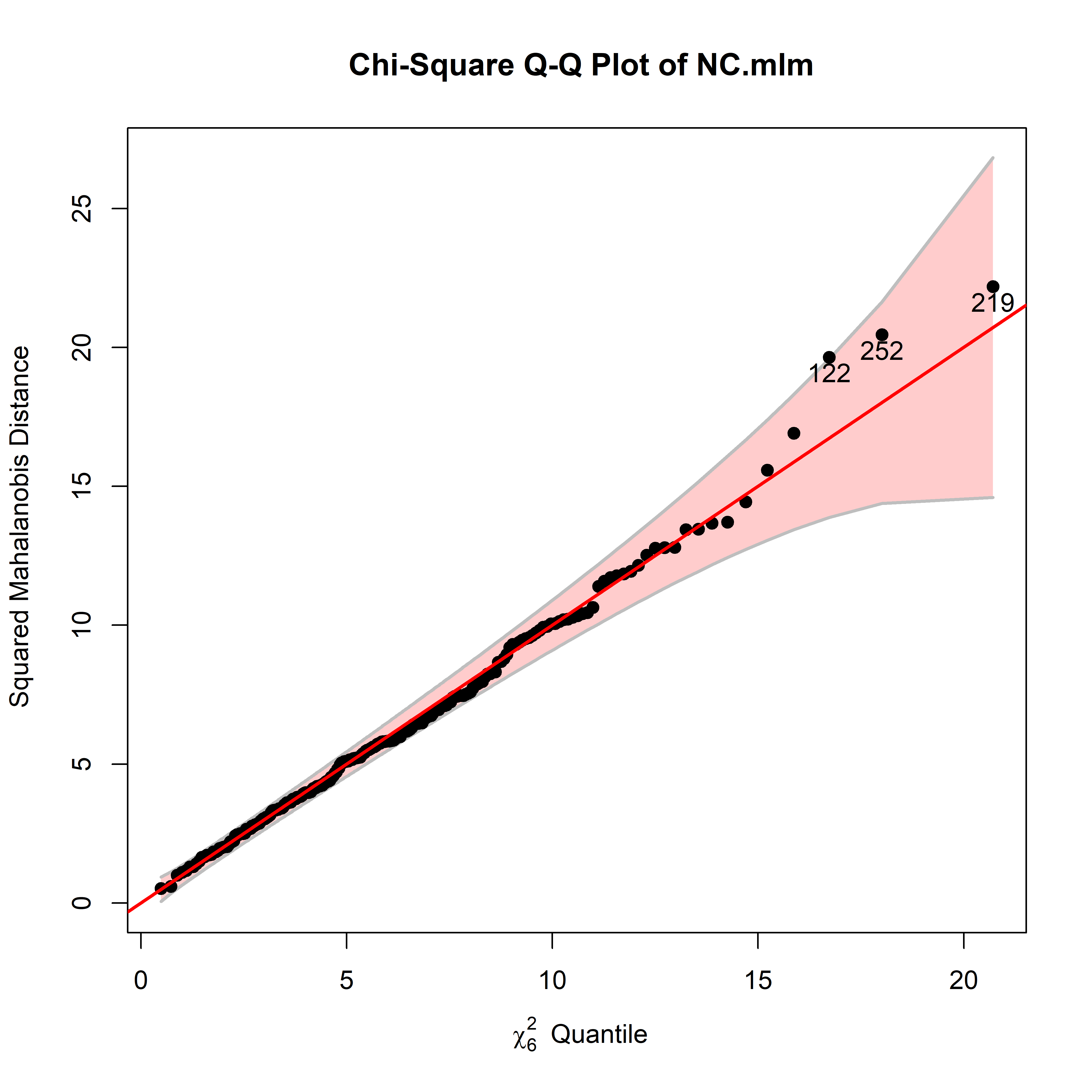
14.3.1 HE plot
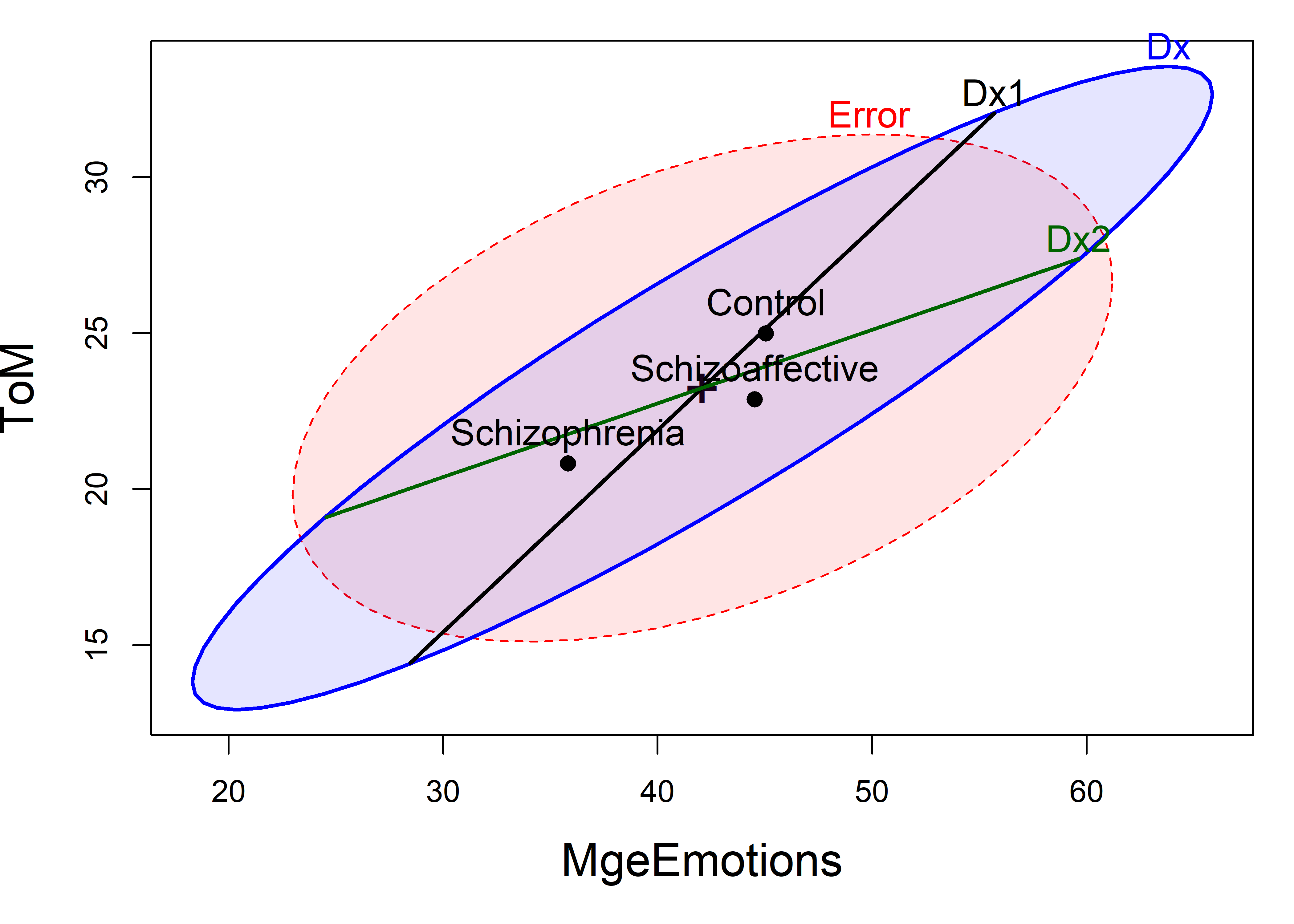
So the question becomes: how to understand these results.heplot() shows the visualization of the multivariate model in the space of two response variables (the first two by default). The result (Figure fig-NC-HEplot) tells a very simple story: The control group performs higher on higher measures than the diagnosed groups, which do not differ between themselves.
(Because heplot() gets the levels of factors from the model object, in order to abbreviate the group labels in the plot, you need to update() the MLM model after the labels are reassigned.)
Then, feed the model to heplot() for a plot of the first two response variables, Speed and Attention.
heplot(NC.mlm,
fill=TRUE, fill.alpha=0.1,
cex.lab=1.5, cex=1.35)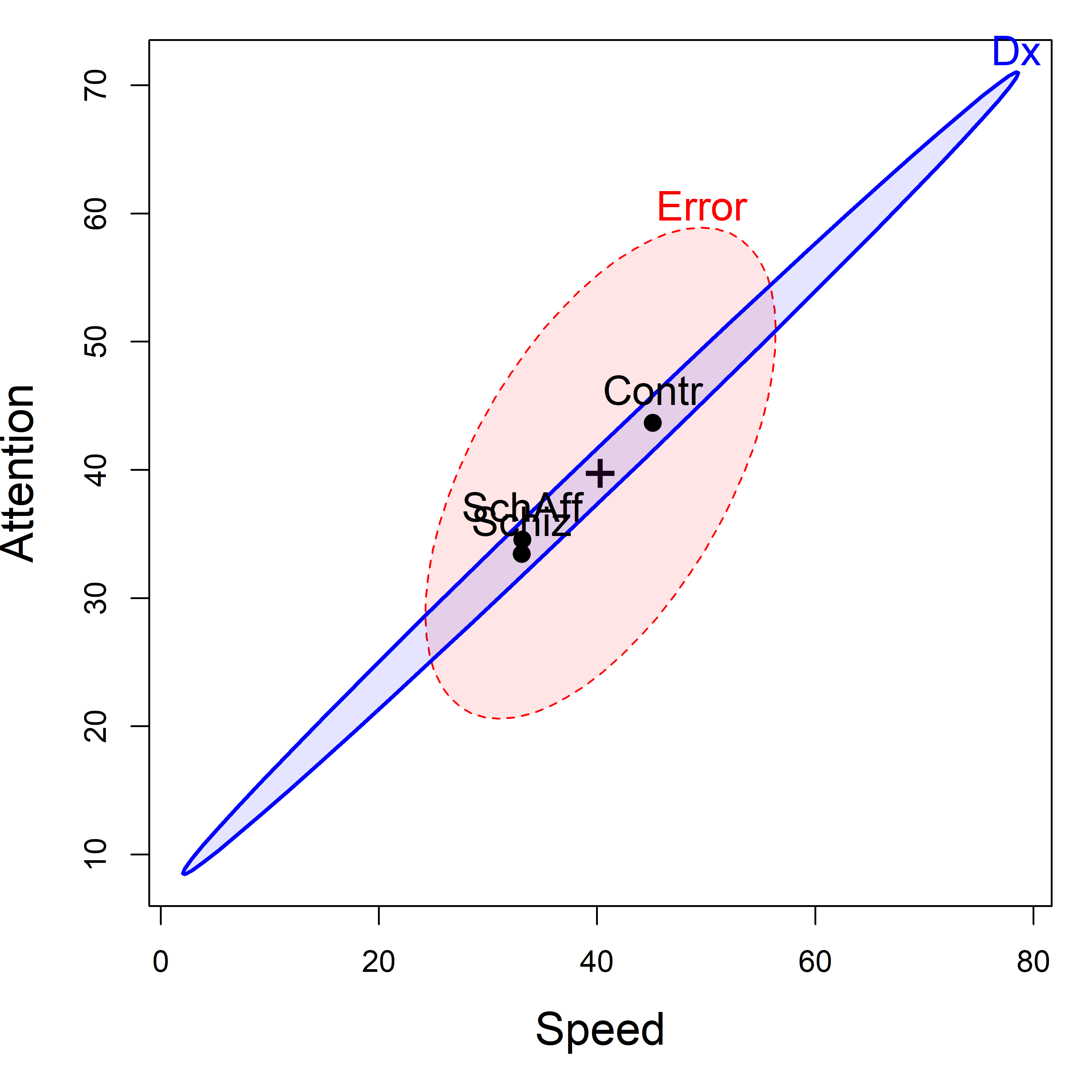
NeuroCog data. The labeled points show the means of the groups on the two variables. The blue \(\mathbf{H}\) ellipse for groups indicates the strong positive correlation of the group means.
This pattern, of the control group higher than the others, is consistent across all of the response variables, as we see from a plot of pairs(NC.mlm) in Figure fig-NC-HE-pairs.
pairs(NC.mlm,
fill=TRUE, fill.alpha=0.1,
var.cex=2)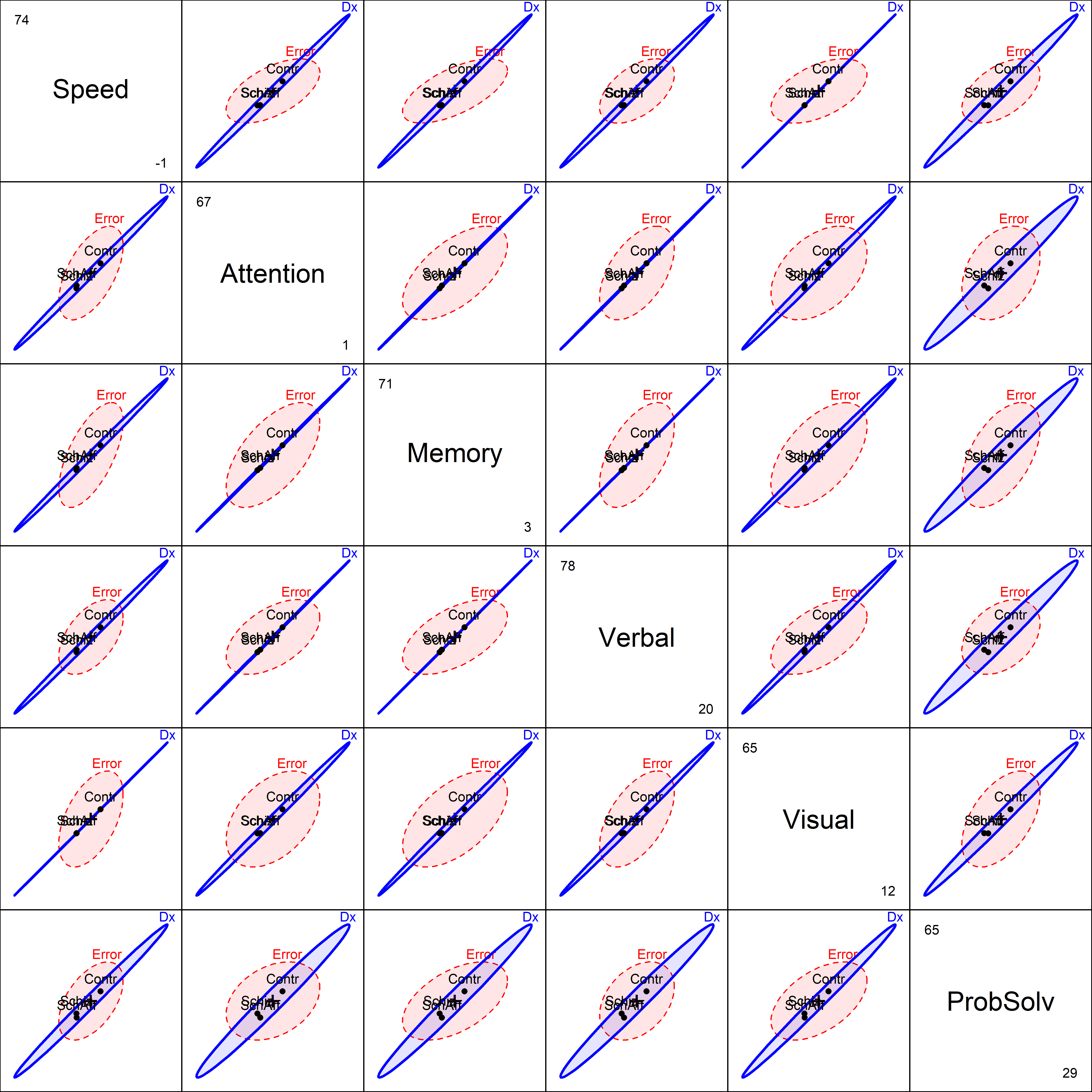
NeuroCog data. The visual evidence that these outcome measures are strongly related in distinguishing the groups is very clear.
The group means are nearly perfectly correlated in all panels. This signals that we are likely to see a very simple representation of the data in canonical space. Note that this differs from the biplot view (Figure fig-neuro-biplot) in that biplot ignores the Dx factor in the PCA, even though we represent it in the plot.
14.3.2 Canonical space
We can gain further insight, and a simplified plot showing all the response variables by projecting the MANOVA into the canonical space, which is entirely 2-dimensional (because \(\text{df}_h=2\)). However, the output from candisc() shows that 98.5% of the mean differences among groups can be accounted for in just one canonical dimension.
NC.can <- candisc(NC.mlm)
NC.can
#
# Canonical Discriminant Analysis for Dx:
#
# CanRsq Eigenvalue Difference Percent Cumulative
# 1 0.29295 0.41433 0.408 98.5 98.5
# 2 0.00625 0.00629 0.408 1.5 100.0
#
# Test of H0: The canonical correlations in the
# current row and all that follow are zero
#
# LR test stat approx F numDF denDF Pr(> F)
# 1 0.703 7.53 12 468 9e-13 ***
# 2 0.994 0.30 5 235 0.91
# ---
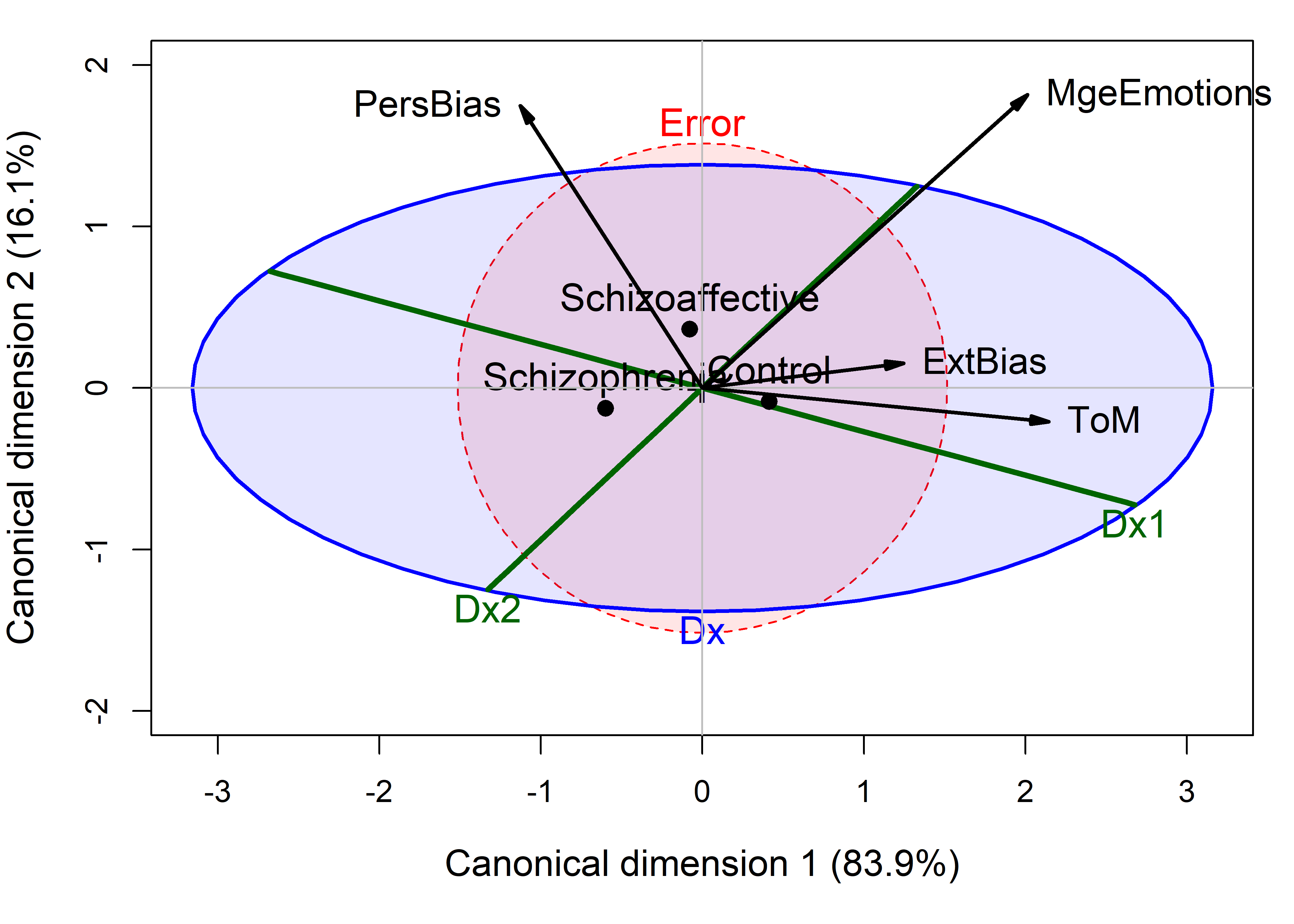
# Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Figure fig-NC-candisc is the result of the plot() method for class "candisc" objects, that is, the result of calling plot(NC.can, ...). It plots the two canonical scores, \(\mathbf{Z}_{n \times 2}\) for the subjects, together with data ellipses for each of the three groups.
pos <- c(4, 1, 4, 4, 1, 3)
col <- c("red", "darkgreen", "blue")
plot(NC.can,
ellipse=TRUE,
rev.axes=c(TRUE,FALSE),
pch=c(7,9,10),
var.cex=1.2, cex.lab=1.5, var.lwd=2, scale=4.5,
col=col,
var.col="black", var.pos=pos,
prefix="Canonical dimension ")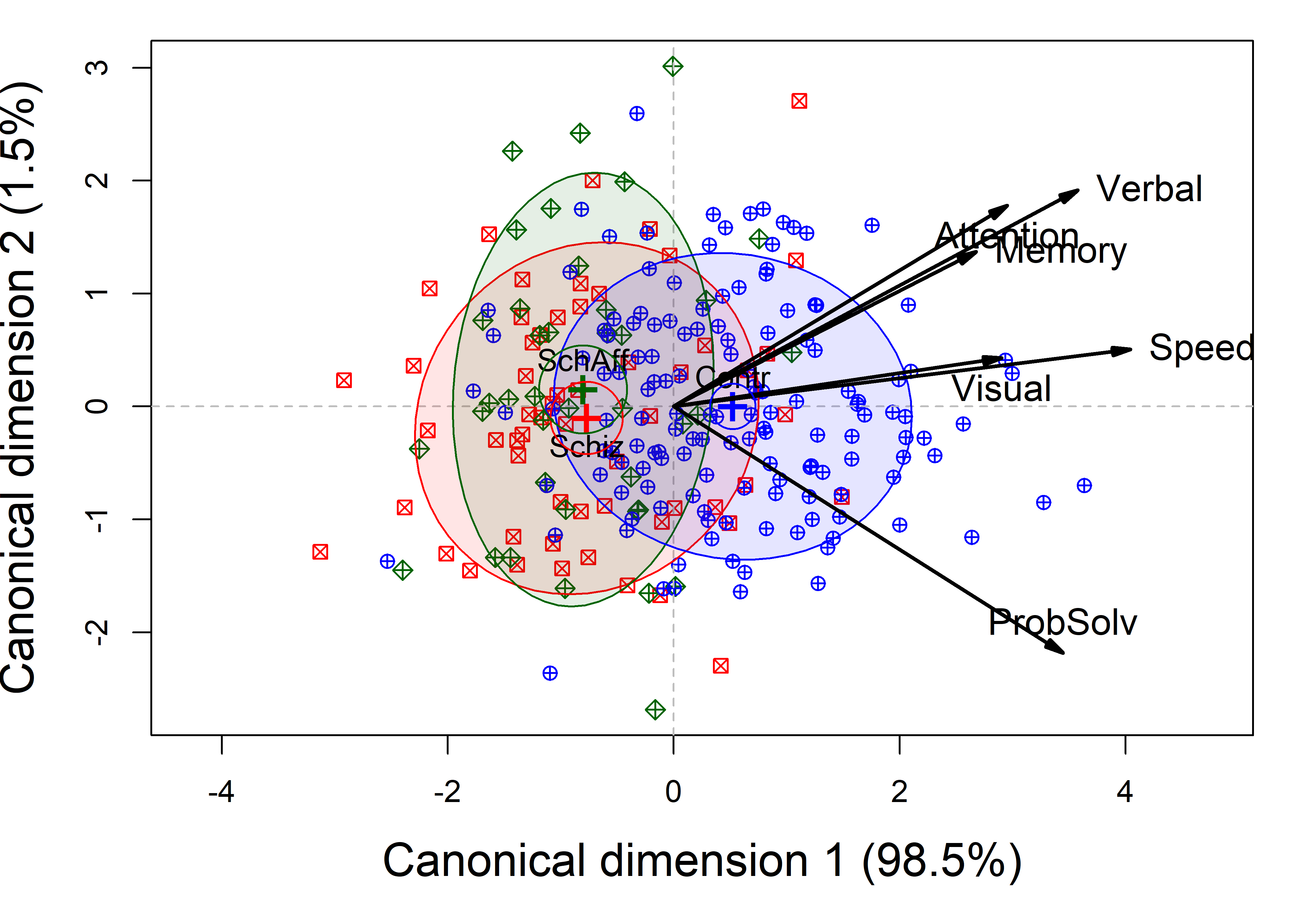
NeuroCog data MANOVA. Scores on the two canonical dimensions are plotted, together with 68% data ellipses for each group.
The interpretation of Figure fig-NC-candisc is again fairly straightforward. As noted earlier (sec-candisc), the projections of the variable vectors in this plot on the coordinate axes are proportional to the correlations of the responses with the canonical scores. From this, we see that the normal group differs from the two patient groups, having higher scores on all the neurocognitive variables, most of which are highyl correlated. The problem solving measure is slightly different, and this, compared to the cluster of memory, verbal and attention, is what distinguishes the schizophrenic group from the schizoaffectives.
The separation of the groups is essentially one-dimensional, with the control group higher on all measures. Moreover, the variables processing speed and visual memory are the purest measures of this dimension, but all variables contribute positively. The second canonical dimension accounts for only 1.5% of group mean differences and is non-significant (by a likelihood ratio test). Yet, if we were to interpret it, we would note that the schizophrenia group is slightly higher on this dimension, scoring better in problem solving and slightly worse on working memory, attention, and verbal learning tasks.
Summary
This analysis gives a very simple description of the data, in relation to the research questions posed earlier:
On the basis of these neurocognitive tests, the schizophrenic and schizoaffective groups do not differ significantly overall, but these groups differ greatly from the normal controls.
All cognitive domains distinguish the groups in the same direction, with the greatest differences shown for the variables most closely aligned with the horizontal axis in Figure fig-NC-candisc. This means that these neuro-psychological measures can be considered to tap a single dimension of differences among the three diagnostic groups.
More worked out examples are contained in the heplots vignettes
vignette("HE_manova", package="heplots")andvignette("HE_mmra", package="heplots")↩︎Actually, multivariate normality of the predictors in \(\mathbf{X}\) is not required in the MLM. This assumption applies only to the conditional values \(\mathbf{Y} \;|\; \mathbf{X}\), i.e., that the errors \(\boldsymbol{\epsilon}_{i}' \sim \mathcal{N}_{p}(\mathbf{0},\boldsymbol{\Sigma})\) with constant covariance matrix. Moreover, the widely used MVN test statistics, such as Mardia’s (1970) test based on multivariate skewness and kurtosis are known to be quite sensitive to mild departures in kurtosis (Mardia, 1974) which do not threaten the validity of the multivariate tests.↩︎
The direct application of significance tests to canonical scores probably requires some adjustment because these are computed to have the optimal between-group discrimination.↩︎